When you buy through linkup on our site , we may clear an affiliate commission . Here ’s how it work .
Anartificial intelligence(AI ) model has feign half a billion days of molecular evolution to create the code for a previously obscure protein , according to a new study . The glowing protein , which is similar to those found in man-of-war and red coral , may serve in the development of newfangled medical specialty , researchers say .
Proteins are one of the building block of life and do various functions in the body , such asbuilding musclesand fighting disease . The simulated protein , name esmGFP , only exists as figurer code , but contains the blueprint for a previously obscure type of green fluorescent protein . In nature , green fluorescent proteins give fluorescent Portuguese man-of-war and corals their gleam .
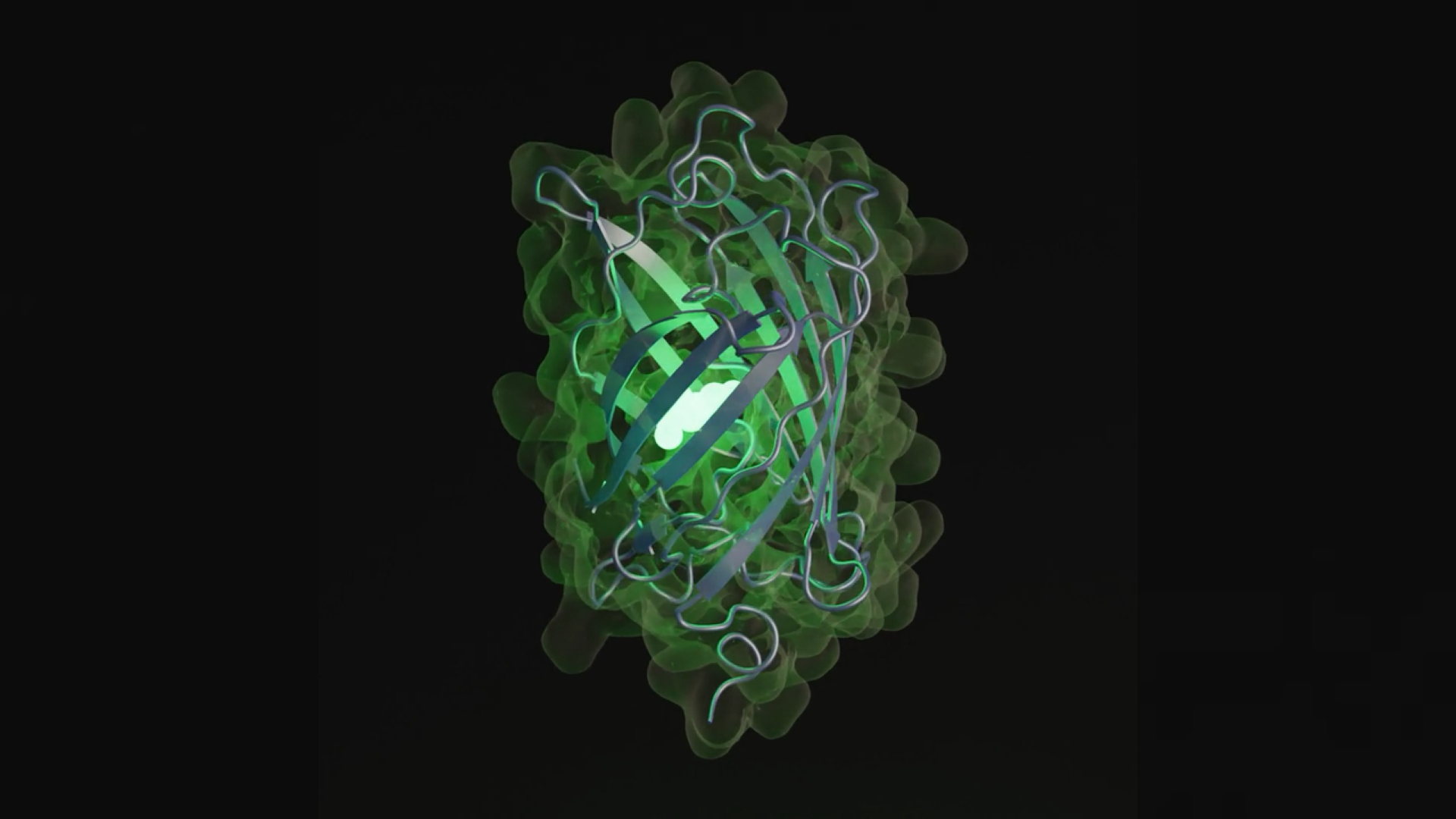
An artist’s depiction of esmGFP, the new fluorescent protein created by ESM3.
The sequence of letter that spell out the education to make esmGFP is only 58 % similar to the closest known fluorescent protein , which is a human - modify interpretation of a protein find in house of cards - tip sea anemones ( Entacmaea quadricolor ) — colorful sea wight that await like they have bubble on the terminal of their tentacles . The relaxation of the sequence is unique , and would require a total of 96 different genetic mutation to germinate . These change would have taken more than 500 million years to evolve of course , harmonize to the survey .
Researchers at a fellowship calledEvolutionaryScaleunveiled esmGFPand the AI model used to create it , ESM3 , in a preprint study last year . self-governing scientists have now peer - review those determination , which were published Jan. 16 in the journalScience .
ESM3 does n’t contrive protein within the common constraints of evolution . Instead , it ’s a problem - solver that fill in gaps of incomplete protein codification put up by the researchers , and in doing so designs something that could exist based on all of the likely pathways development could take .
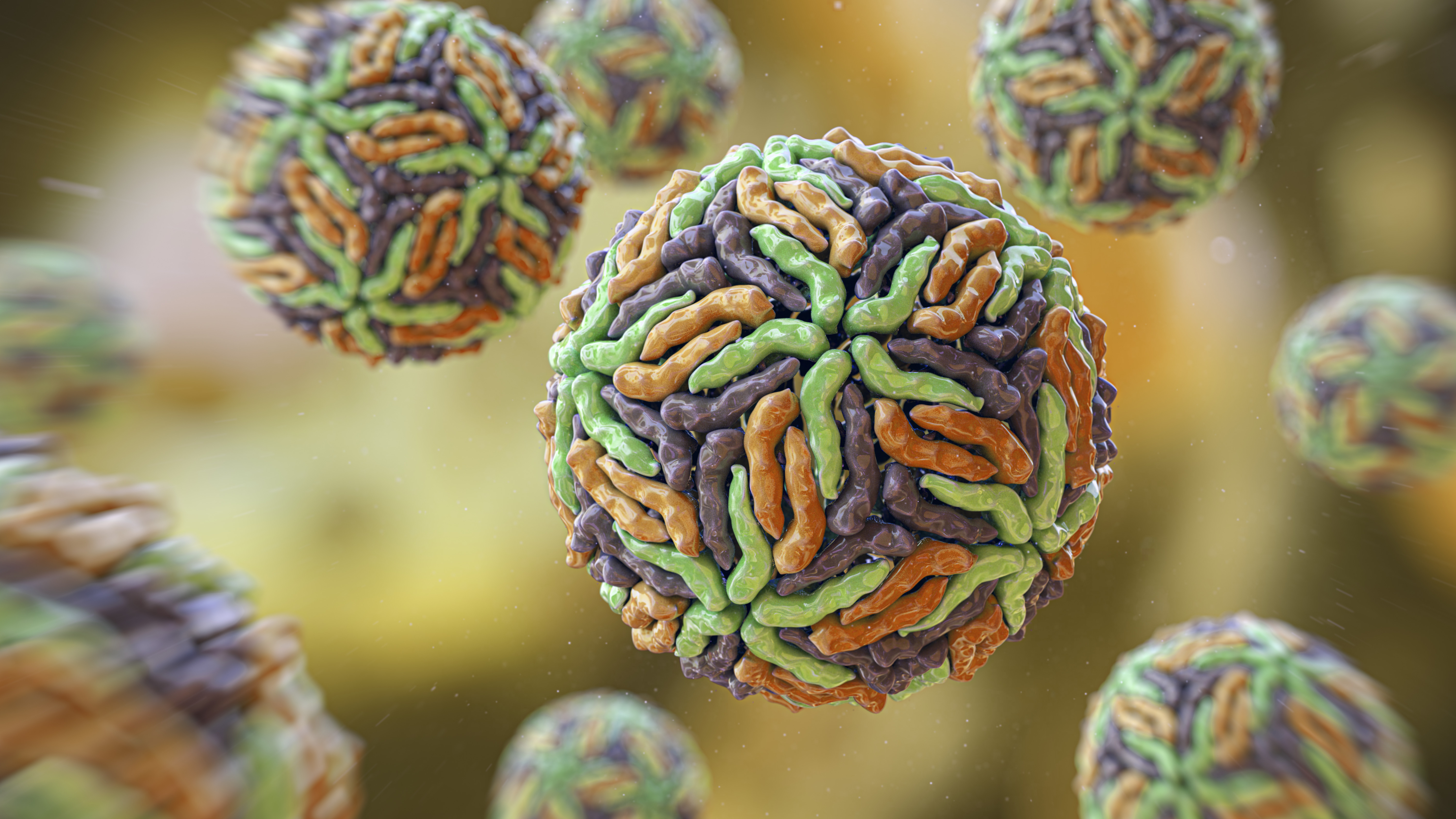
" We ’ve found that ESM3 learns fundamental biota , and can generate functional protein outside the space search by evolution , " field cobalt - authorAlex Rives , co - beginner and chief scientist of EvolutionaryScale , told Live Science in an email .
Related : Taiwanese researchers just built an undetermined - source rival to ChatGPT in 2 months . Silicon Valley is freaked out .
The novel sketch builds on research that Rives and his colleaguesbegan at Meta , the parent company of Facebook and Instagram , before start EvolutionaryScale in 2024 . ESM3 is their late version of a procreative language model similar to OpenAI ’s GPT-4 , which feed ChatGPT , but it ’s based on biology .

Proteins are made up of chain of particle called amino group battery-acid , the sequence of which is provided by genes . Different proteins have unlike amino group battery-acid successiveness . They also disagree structurally , each folding into a unique shape that allow them to carry out their function , according toNature Education . For ESM3 to empathise proteins , researchers feed the theoretical account datum on the main property of a protein — amino acid sequence , anatomical structure and single-valued function — as a series of letters .
The team train ESM3 on data from 2.78 billion proteins found in nature . The researchers then randomly obscure portion of a protein pattern and had ESM3 plug in the gaps to complete the code establish on what it had learned .
" The same way a person can fill in the blanks in the soliloquy " to _ or not to _ , that is the _ , " we can train a linguistic communication model to fill in the blank in proteins , " Rives allege . " Our research has shown that by puzzle out this simple-minded task , information about the deep structure of protein biology emerge in the meshwork . "

— scientist propose take AI suffer to see if it ’s sentient
— AI can now reduplicate itself — a milestone that has experts terrified
— AI could crack unsolvable problems — and humans wo n’t be able to realise the resultant role
Scientists already modify natural proteins and organise new ones for a diversity of aim . For example , greenish fluorescent proteins are used widely in inquiry labs . Their genetic code is often add to the ending of other DNA sequence to sprain the proteins that they encode green . This allows scientist to easily cover protein and cellular processes . Rives noted that ESM3 ’s capabilities can accelerate a broad range of a function of diligence for protein engineering , let in with aid to plan new drug .
Tiffany Taylor , an evolutionary biologist at the University of Bath in the U.K. who was not involved in the research , report on the preprint version of the subject for Live Science in 2024 . In her depth psychology , Taylor wrote that AI models like ESM3 will enable innovations in protein engineering that evolution ca n’t . However , she also noted that the researcher ' claim of assume 500 million yr of evolution is focussed only on single proteins and does not describe for the many stage of natural selection that ultimately create life sentence .
" AI - driven protein engine room is intriguing , but I ca n’t facilitate feel we might be overly confident in assuming we can outfox the intricate cognitive operation honed by millions of years of rude selection , " Taylor said .

You must confirm your public display name before commenting
Please logout and then login again , you will then be prompted to enter your display name .